What do I need to consider when measuring the zeta potential of biological samples?
Size and zeta potential
TRPS is perfectly suited to measure the zeta potential of a range of biological samples, ranging from EVs, liposomes, viruses to DNA modified particles.
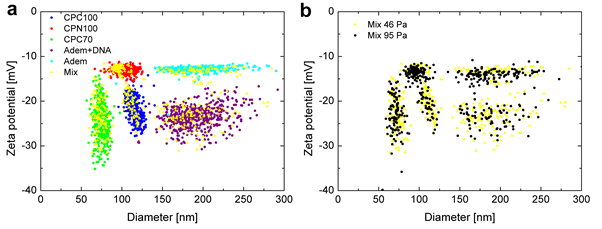
In below figure, an example of a penta-modal mix of five particles, including bare polystyrene (CPN100), carboxylated polystyrene (CPC70, CPC100), unmodified and DNA-functionalised magnetic nanoparticles (Adem) is shown, that could be fully resolved in both size and zeta potential with TRPS. The associated distributions are not affected by different applied pressures, confirming reproducibility and robustness of this TRPS protocol.
Kinetic Study
Highly sensitive kinetic studies can be carried out with TRPS.
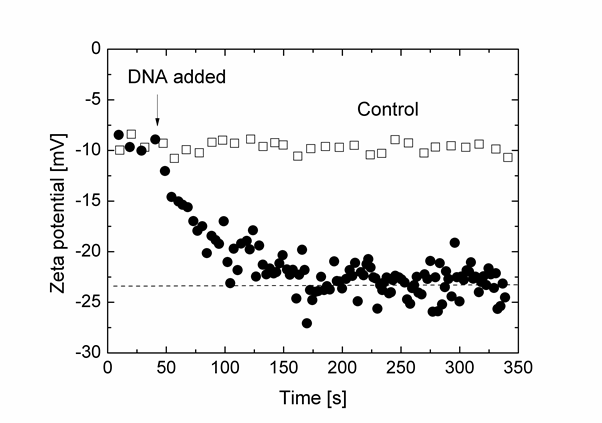
An example is shown that demonstrates the immobilisation of oligonucleotides onto magnetic particles, providing information about particle properties that is of utmost importance for applications like DNA sequencing, bio-sensing and drug delivery. The graphics shows zeta potential measurements overtime, illustrating the binding capacity of surface-functionalised magnetic particles to DNA. Compared with a control sample, immobilisation of DNA onto the surface of the streptavidin-coated superparamagnetic particles was seen to complete in approximately 100 seconds after the addition of the bio-targets.
Care needs to be taken when analysing complex biological samples to prevent particle-pore interactions and surface modification of the pore.
For most biological samples nanopore coating solution must be used to alleviate non-specific binding to the pore surface.
It is good practice to carry out additional runs using external calibrants at the end of zeta potential measurements to confirm that the tunable pore remains intact and unchanged during analysis.